Construct and Train the Network with Multiple Outputs in MATLAB
Introduction
The MATLAB official example, “Train Network with Multiple Outputs ” 1, shows how to construct a multi-output neural network using custom training loop.
Actually, as mentioned in the official documentation, “Multiple-Input and Multiple-Output Networks”2, if the constructed network has multiple outputs, using custom training loop is the only choice, that is
trainNetworkfunction is not available:
It is a good example shows how to construct neural networks to solve multi-label problems in MATLAB. Therefore, I would record it down in this post and make some notes about it.
Reproduce the example: train and test the network
Specifically, this example is to show “how to train a deep learning network with multiple outputs that predict both labels and angles of rotations of handwritten digits”1. And the necessary code for preparing training data set, constructing and training the network shows as follows:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
clc,clear,close all
rng("default")
% Load training data
[XTrain,T1Train,T2Train] = digitTrain4DArrayData;
dsXTrain = arrayDatastore(XTrain,IterationDimension=4);
dsT1Train = arrayDatastore(T1Train);
dsT2Train = arrayDatastore(T2Train);
dsTrain = combine(dsXTrain,dsT1Train,dsT2Train);
classNames = categories(T1Train);
numClasses = numel(classNames);
numObservations = numel(T1Train);
% Define deep learning model
% Define the main block of layers as a layer graph.
layers = [
imageInputLayer([28 28 1],Normalization="none")
convolution2dLayer(5,16,Padding="same")
batchNormalizationLayer
reluLayer(Name="relu_1")
convolution2dLayer(3,32,Padding="same",Stride=2)
batchNormalizationLayer
reluLayer
convolution2dLayer(3,32,Padding="same")
batchNormalizationLayer
reluLayer
additionLayer(2,Name="add")
fullyConnectedLayer(numClasses)
softmaxLayer(Name="softmax")];
lgraph = layerGraph(layers);
% And the skip connection
layers = [
convolution2dLayer(1,32,Stride=2,Name="conv_skip")
batchNormalizationLayer
reluLayer(Name="relu_skip")];
lgraph = addLayers(lgraph,layers);
lgraph = connectLayers(lgraph,"relu_1","conv_skip");
lgraph = connectLayers(lgraph,"relu_skip","add/in2");
% Add the fully connected layer for regression.
layers = fullyConnectedLayer(1,Name="fc_2");
lgraph = addLayers(lgraph,layers);
lgraph = connectLayers(lgraph,"add","fc_2");
% Create a dlnetwork object from the layer graph.
net = dlnetwork(lgraph);
% Specify Training Options
numEpochs = 30;
miniBatchSize = 128;
% Train the network
mbq = minibatchqueue(dsTrain,...
MiniBatchSize=miniBatchSize,...
MiniBatchFcn=@preprocessData,...
MiniBatchFormat=["SSCB" "" ""]);
% Initialize the training progress plot.
figure("Color","w")
hold(gca,"on"),box(gca,"on"),grid(gca,"on")
C = colororder;
lineLossTrain = animatedline("Color",C(2,:));
ylim([0 inf])
xlabel("Iteration")
ylabel("Loss")
% Initialize parameters for Adam.
trailingAvg = [];
trailingAvgSq = [];
% Train the model
iteration = 0;
start = tic;
% Loop over epochs.
for epoch = 1:numEpochs
% Shuffle data.
shuffle(mbq)
% Loop over mini-batches
while hasdata(mbq)
iteration = iteration + 1;
[X,T1,T2] = next(mbq);
% Evaluate the model loss, gradients, and state using
% the dlfeval and modelLoss functions.
[loss,gradients,state] = dlfeval(@modelLoss,net,X,T1,T2);
net.State = state;
% Update the network parameters using the Adam optimizer.
[net,trailingAvg,trailingAvgSq] = adamupdate(net,gradients, ...
trailingAvg,trailingAvgSq,iteration);
% Display the training progress.
D = duration(0,0,toc(start),Format="hh:mm:ss");
loss = double(loss);
addpoints(lineLossTrain,iteration,loss)
title("Epoch: " + epoch + ", Elapsed: " + string(D))
drawnow
end
end
% Copyright 2019 The MathWorks, Inc.
where model loss function modelLoss and mini-batch preprocessing function preprocessData are respectively:
1
2
3
4
5
6
7
8
9
10
% Define model loss function
function [loss,gradients,state] = modelLoss(net,X,T1,T2)
[Y1,Y2,state] = forward(net,X,Outputs=["softmax" "fc_2"]);
lossLabels = crossentropy(Y1,T1);
lossAngles = mse(Y2,T2);
loss = lossLabels + 0.1*lossAngles;
gradients = dlgradient(loss,net.Learnables);
end
1
2
3
4
5
6
7
8
9
10
11
12
13
14
% Define mini-batch preprocessing function
function [X,T1,T2] = preprocessData(dataX,dataT1,dataT2)
% Extract image data from cell and concatenate
X = cat(4,dataX{:});
% Extract label data from cell and concatenate
T1 = cat(2,dataT1{:});
% Extract angle data from cell and concatenate
T2 = cat(2,dataT2{:});
% One-hot encode labels
T1 = onehotencode(T1,1);
end
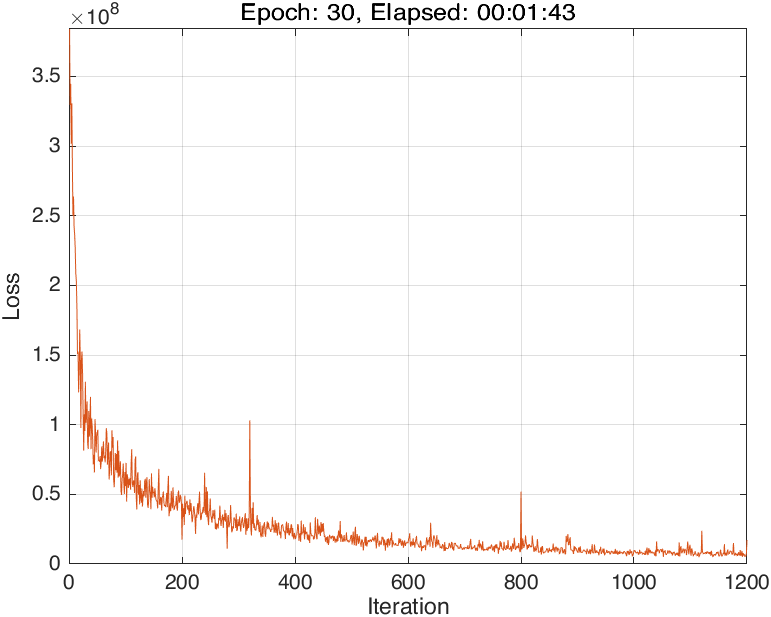
The training progress of running Script 1 looks like:
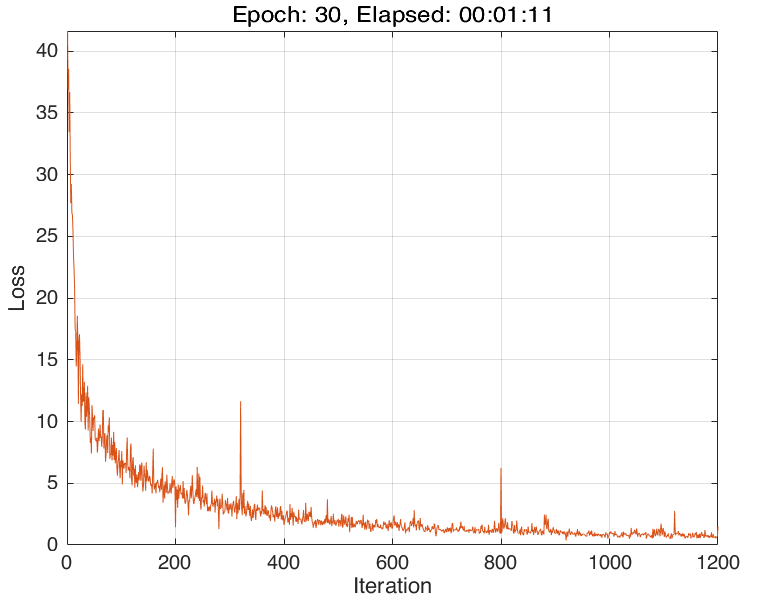
and we could evaluate the well-trained neural network using the following script:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
% Load test data set
[XTest,T1Test,T2Test] = digitTest4DArrayData;
dsXTest = arrayDatastore(XTest,IterationDimension=4);
dsT1Test = arrayDatastore(T1Test);
dsT2Test = arrayDatastore(T2Test);
dsTest = combine(dsXTest,dsT1Test,dsT2Test);
mbqTest = minibatchqueue(dsTest,...
MiniBatchSize=miniBatchSize,...
MiniBatchFcn=@preprocessData,...
MiniBatchFormat=["SSCB" "" ""]);
classesPredictions = [];
anglesPredictions = [];
classCorr = [];
angleDiff = [];
% Loop over mini-batches.
while hasdata(mbqTest)
% Read mini-batch of data.
[X,T1,T2] = next(mbqTest);
% Make predictions using the predict function.
[Y1,Y2] = predict(net,X,Outputs=["softmax" "fc_2"]);
% Determine predicted classes.
Y1 = onehotdecode(Y1,classNames,1);
classesPredictions = [classesPredictions Y1]; %#ok
% Dermine predicted angles
Y2 = extractdata(Y2);
anglesPredictions = [anglesPredictions Y2]; %#ok
% Compare predicted and true classes
T1 = onehotdecode(T1,classNames,1);
classCorr = [classCorr Y1 == T1]; %#ok
% Compare predicted and true angles
angleDiffBatch = Y2 - T2;
angleDiffBatch = extractdata(gather(angleDiffBatch));
angleDiff = [angleDiff angleDiffBatch]; %#ok
end
% Evaluate the classification accuracy.
accuracy = mean(classCorr);
% Evaluate the regression accuracy
angleRMSE = sqrt(mean(angleDiff.^2));
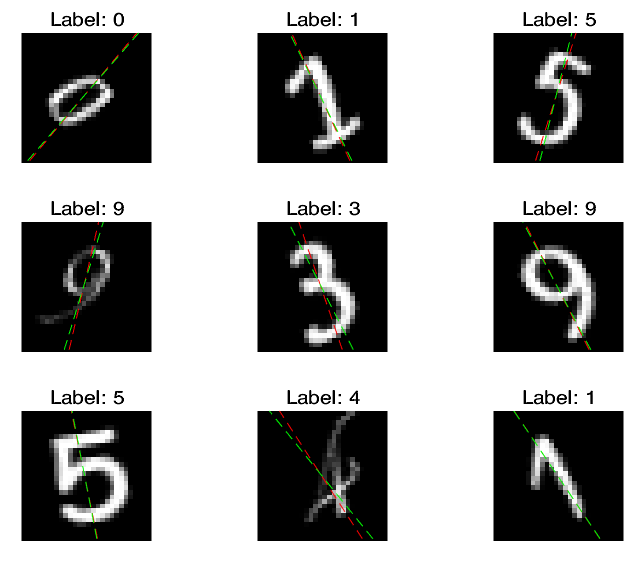
% View some of the images with their predictions.
% Display the predicted angles in red and the correct labels in green.
idx = randperm(size(XTest,4),9);
figure("Color","w")
for i = 1:9
subplot(3,3,i)
I = XTest(:,:,:,idx(i));
imshow(I)
hold on
sz = size(I,1);
offset = sz/2;
thetaPred = anglesPredictions(idx(i));
plot(offset*[1-tand(thetaPred) 1+tand(thetaPred)],[sz 0],"r--")
thetaValidation = T2Test(idx(i));
plot(offset*[1-tand(thetaValidation) 1+tand(thetaValidation)],[sz 0],"g--")
hold off
label = string(classesPredictions(idx(i)));
title("Label: " + label)
end
% Copyright 2019 The MathWorks, Inc.
The two measures indicating the model performance, accuracy for classification task and angleRMSE for regression task, are:
1
2
3
4
5
6
7
>> accuracy, angleRMSE
accuracy =
0.9792
angleRMSE =
single
6.8784
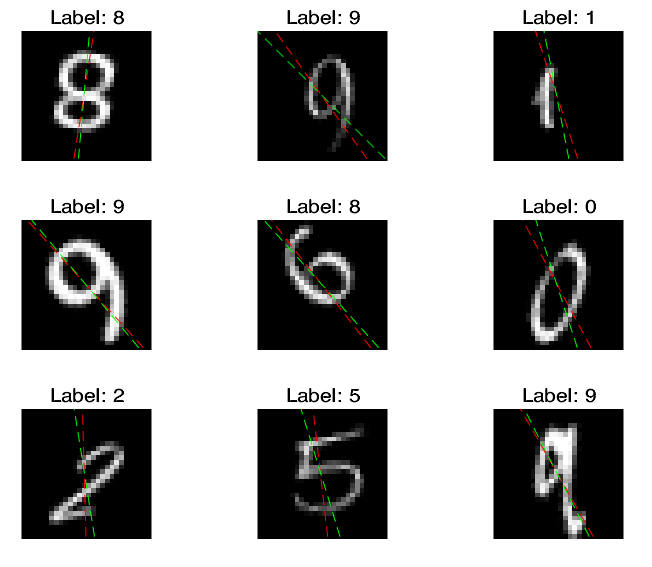
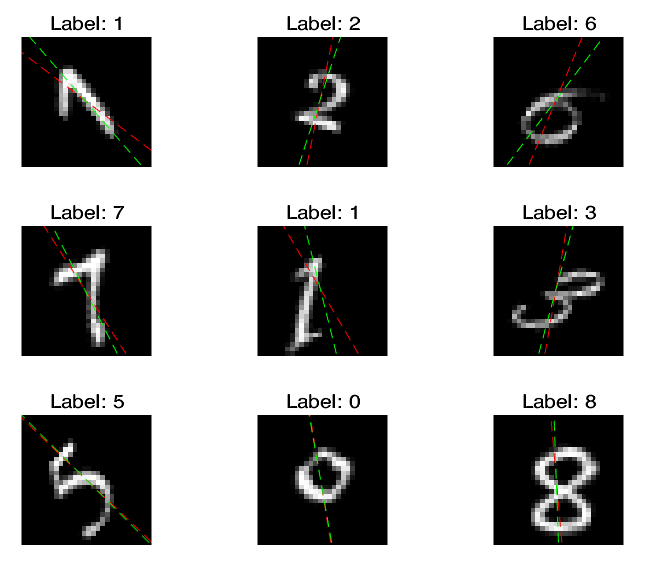
where the green lines represent the true angles, and red lines represent the predicted angles.
Discussions
Prepare data sets
In Script 1, we use the following code to prepare training data set for training the network (so do for preparing test data set in Script 2):
1
2
3
4
5
6
7
8
% Load training data
[XTrain,T1Train,T2Train] = digitTrain4DArrayData;
dsXTrain = arrayDatastore(XTrain,IterationDimension=4);
dsT1Train = arrayDatastore(T1Train);
dsT2Train = arrayDatastore(T2Train);
dsTrain = combine(dsXTrain,dsT1Train,dsT2Train);
where the data type of XTrain (greyscale image), T1Train (labels for classification), T2Train (labels for regression) is clear:
1
2
3
4
5
6
>> whos XTrain T1Train T2Train
Name Size Bytes Class Attributes
T1Train 5000x1 6062 categorical
T2Train 5000x1 40000 double
XTrain 28x28x1x5000 31360000 double
After importing them, we should use arrayDatastore function3 and combine function4 to prepare data set for training:
1
2
3
4
5
dsXTrain = arrayDatastore(XTrain,IterationDimension=4);
dsT1Train = arrayDatastore(T1Train);
dsT2Train = arrayDatastore(T2Train);
dsTrain = combine(dsXTrain,dsT1Train,dsT2Train);
The default value of IterationDimension property of arrayDatastore function is 1. Easily speaking, the value of IterationDimension property specifies which dimension MATLAB should count the sample size along.
By the way, data actually aren’t stored in variables dsXTrain, dsT1Train, and dsT2Train:
1
2
3
4
5
6
>> whos dsXTrain dsT1Train dsT2Train
Name Size Bytes Class Attributes
dsT1Train 1x1 8 matlab.io.datastore.ArrayDatastore
dsT2Train 1x1 8 matlab.io.datastore.ArrayDatastore
dsXTrain 1x1 8 matlab.io.datastore.ArrayDatastore
arrayDatastore seemingly just defines how to read data from in-memory variables XTrain, T1Train, and T2Train3. And so do for dsTrain, which is created by combine function4:
1
2
3
4
>> whos dsTrain
Name Size Bytes Class Attributes
dsTrain 1x1 8 matlab.io.datastore.CombinedDatastore
To sum up, the whole preparation work is showed as above, and we finally could obtain variable dsTrain, which will be put into minibatchqueue function to create mini-batch.
Neural network structure
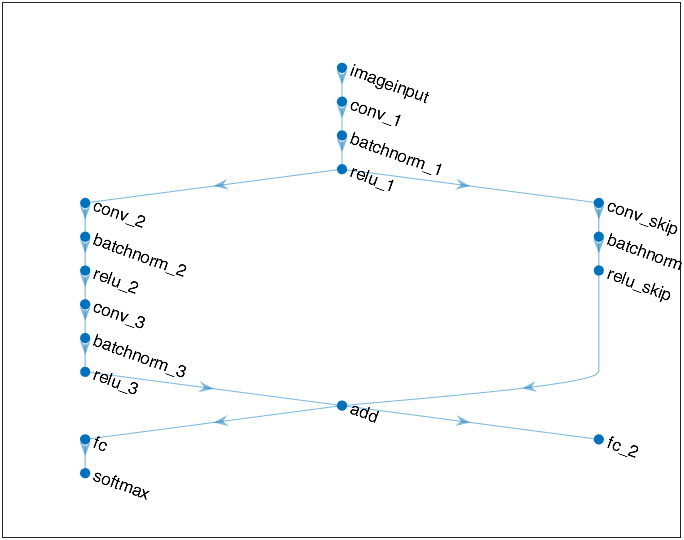
We could use plot(lagraph) or built-in function analyzeNetwork5 to inspect the neural network structure:
1
plot(lgraph)
1
analyzeNetwork(net)
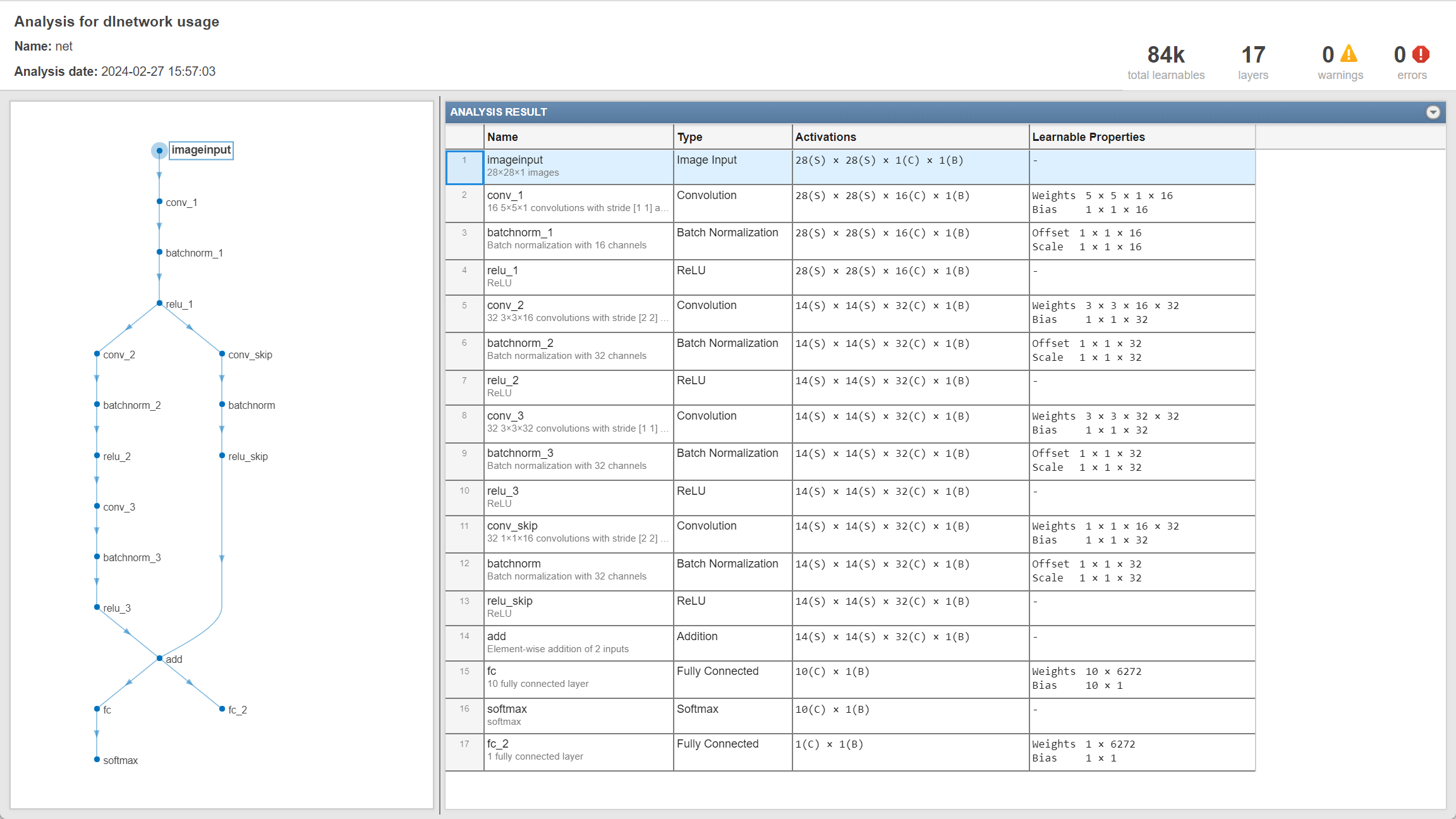
As can be seen, when creating the main block of layers in Script 1, an addition layer is created by additionLayer function6 before creating the last fully-connected layer. Then, a skip connection is then connected to the main branch by addLayers7and connectLayers8 (In fact, as will be showed in Appendix I, this skip connection is not necessary.)
It should be noted that, if we want to train the neural network using custom training loop, we don’t need to use classificationLayer or regressionLayer to create the output layer as we do when using trainNetwork function to train (as in references 9 and 10, respectively). Instead, using softmaxLayer and fullyConnectedLayer to create the output layers and followed by loss calculation is enough.
Model loss function
As mentioned above, the loss value of neural network is calculated by modelLoss function, and the total loss consists of two parts:
Although the official documentation1 doesn’t explain the function of coefficient $0.1$, I guess it is used to balance two loss value and therefore balance two different tasks (Note that this is just my guess, and in fact, if we expand the gap between them to an extreme situation, the result is still good. See Appendix II.). If we modify modelLoss function to make it output two loss values, we could plot how the loss values change as iteration:
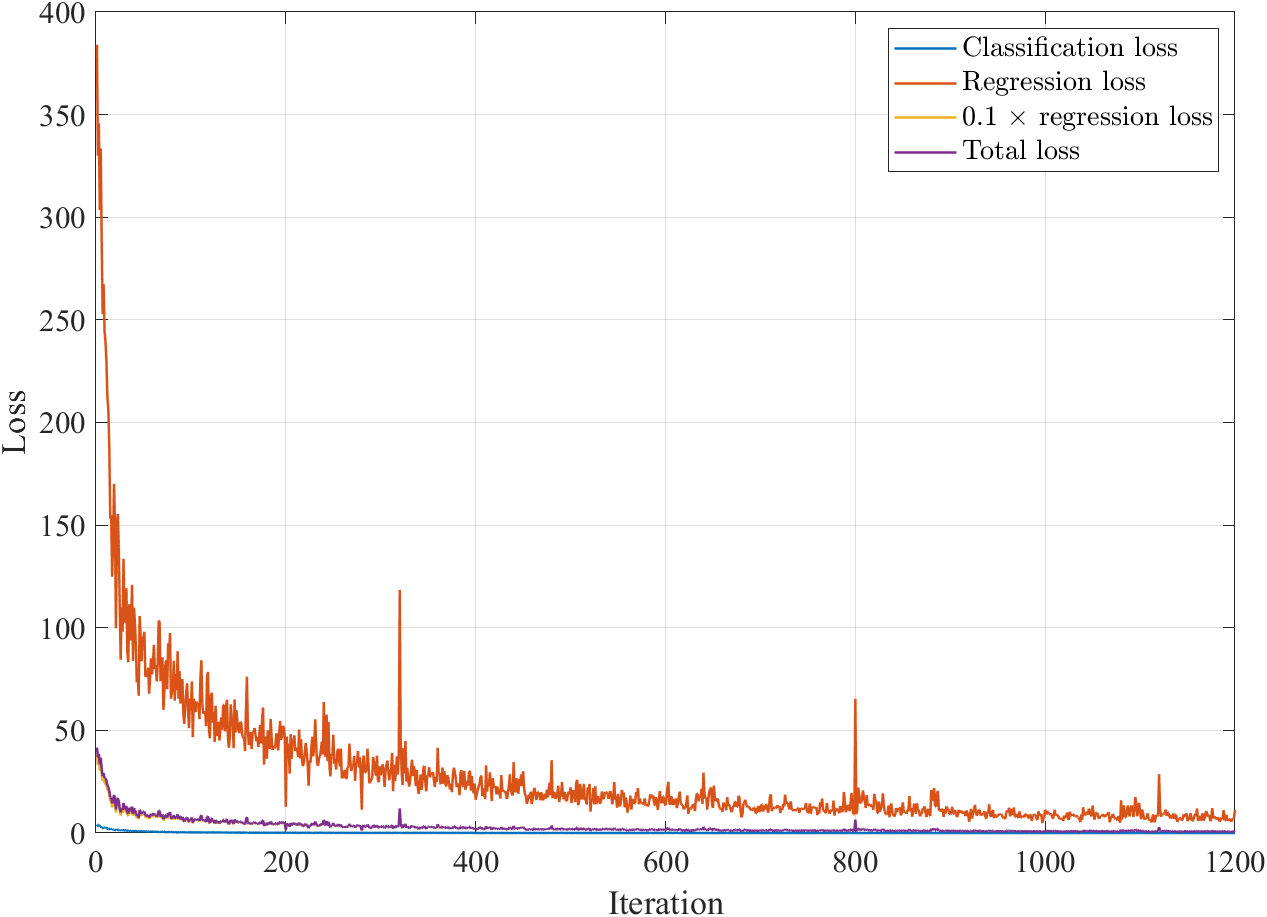
As can be seen, the classification loss, $\text{Cross-entropy}(Y_1,T_1)$, is rather less than the regression regression loss, $\text{MSE}(Y_2,T_2)$, and multiplying $0.1$ before the latter narrows the gap between them.
Appendix
Appendix I
The figure of network architecture may cause a false feeling, that is the main part of the network (the left branch) is for classification, and the right one is for regression:
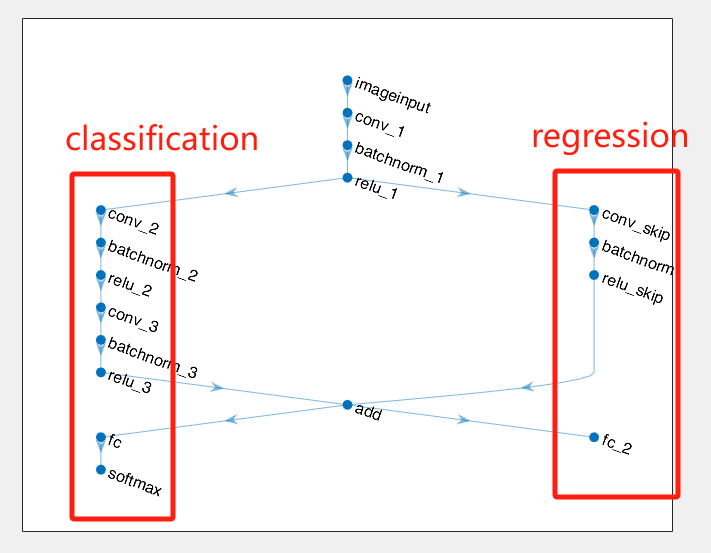
However, it is a wrong understanding! For example, if we delete the right branch, and the network architecture at this time is like:
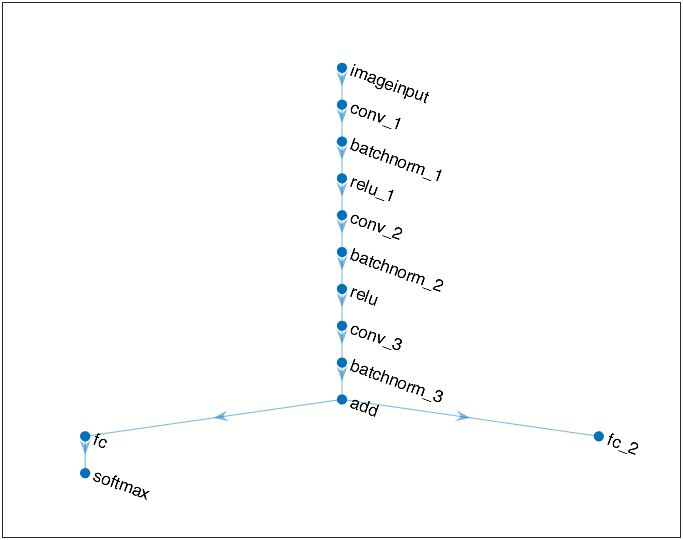
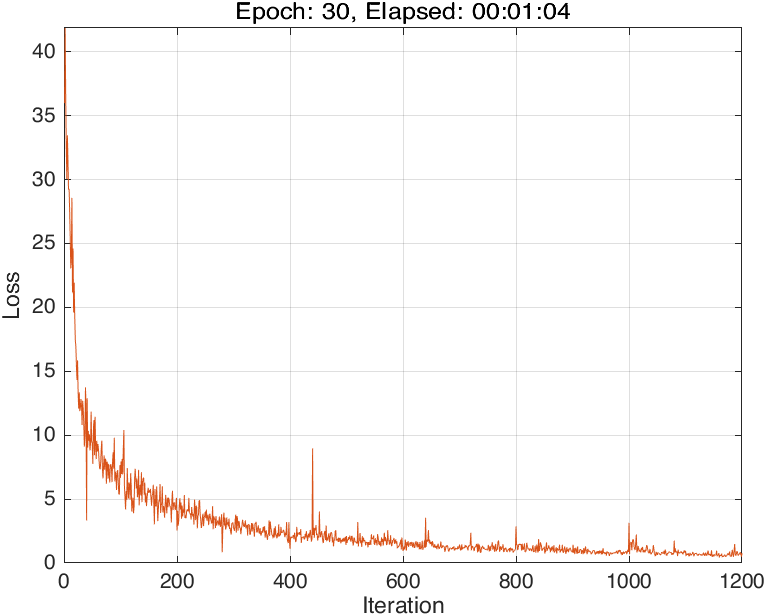
and it also has a good performance:
1
2
3
4
5
6
7
>> accuracy,angleRMSE
accuracy =
0.9810
angleRMSE =
single
6.0480
So, to my mind, the reason a neural network is called multi-output is that it has multiple output layers and different output layers correspond to different loss function calculation methods. In this example, no matter how complex the structure of hidden layers or how many branches it has, in the backward process, the learnable parameters of each layer are optimized by derivating the total loss value which is calculated by $\eqref{eq1}$. In fact, if classification loss and regression loss are derivated for different network branches, then this method is not fundamentally different from establishing two separate neural networks.
Appendix II
Here, I will change the weights of two loss values of modelLoss function to:
that is:
1
2
3
4
5
6
7
8
9
10
% Define the modified model loss function
function [loss,gradients,state,lossLabels,lossAngles] = modelLoss_modified(net,X,T1,T2)
[Y1,Y2,state] = forward(net,X,Outputs=["softmax" "fc_2"]);
lossLabels = crossentropy(Y1,T1);
lossAngles = mse(Y2,T2);
loss = 1e-6*lossLabels + 1e6*lossAngles;
gradients = dlgradient(loss,net.Learnables);
end
In this case, the training progress plot is like:
I expect the classification effect of the network is extremely bad, but the result shows that I am wrong:
1
2
3
4
5
6
7
>> accuracy,angleRMSE
accuracy =
0.9752
angleRMSE =
single
8.8155
Therefore, I am not that sure what the role of coefficient $0.1$ in $\eqref{eq1}$ right now.
References
-
MATLAB
arrayDatastore: Datastore for in-memory data- MathWorks. ˄ ˄2 -
MATLAB
combine: Combine data from multiple datastores - MathWorks. ˄ ˄2 -
MATLAB
analyzeNetwork: Analyze deep learning network architecture - MathWorks. ˄ -
MATLAB
addLayers: Add layers to layer graph or network - MathWorks. ˄ -
MATLAB
connectLayers: Connect layers in layer graph or network - MathWorks. ˄ -
Create Simple Deep Learning Neural Network for Classification - MathWorks. ˄
-
Train Convolutional Neural Network for Regression - MathWorks. ˄
